Pogil Control of Gene Expression in Prokaryotes: The Molecular Mastery Behind Bacterial Precision
Pogil Control of Gene Expression in Prokaryotes: The Molecular Mastery Behind Bacterial Precision
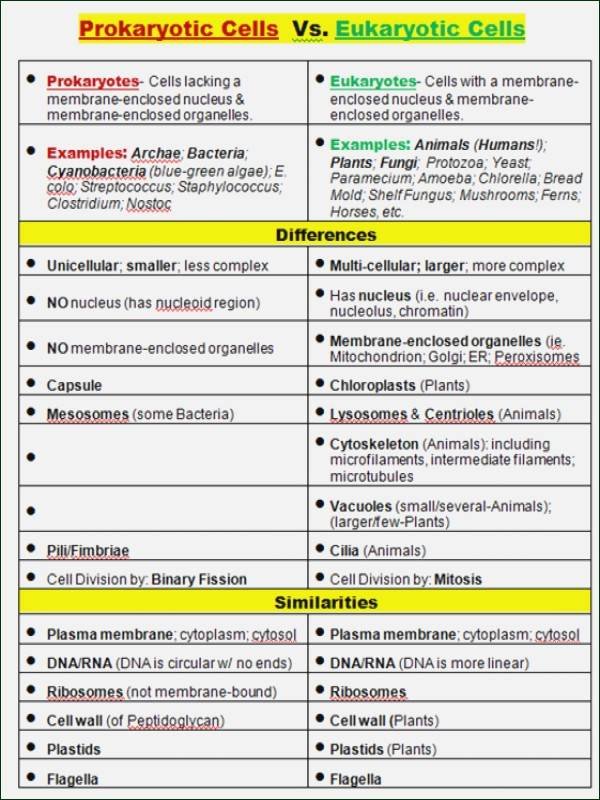
In the silent world of prokaryotes, where microscopic circuits govern survival, the orchestration of gene expression is nothing short of engineering brilliance. Unlike complex eukaryotes, prokaryotic cells rely on compact, highly efficient regulatory mechanisms—core to their rapid adaptation and resilience. Central to this control is the interplay of DNA, RNA, and proteins, all choreographed under the guidance of key regulatory proteins.
The Principles of Gene Expression in Prokaryotes, as explored through the Pogil framework, reveals a masterclass in precision: from transcriptional activation to translational control, every step is tightly regulated to optimize energy and response speed. Understanding how prokaryotes fine-tune their genetic output not only deepens insight into fundamental biology but also fuels innovations in biotechnology and medicine. At the heart of prokaryotic gene regulation lies RNA polymerase, the molecular engine responsible for transcribing DNA into RNA.
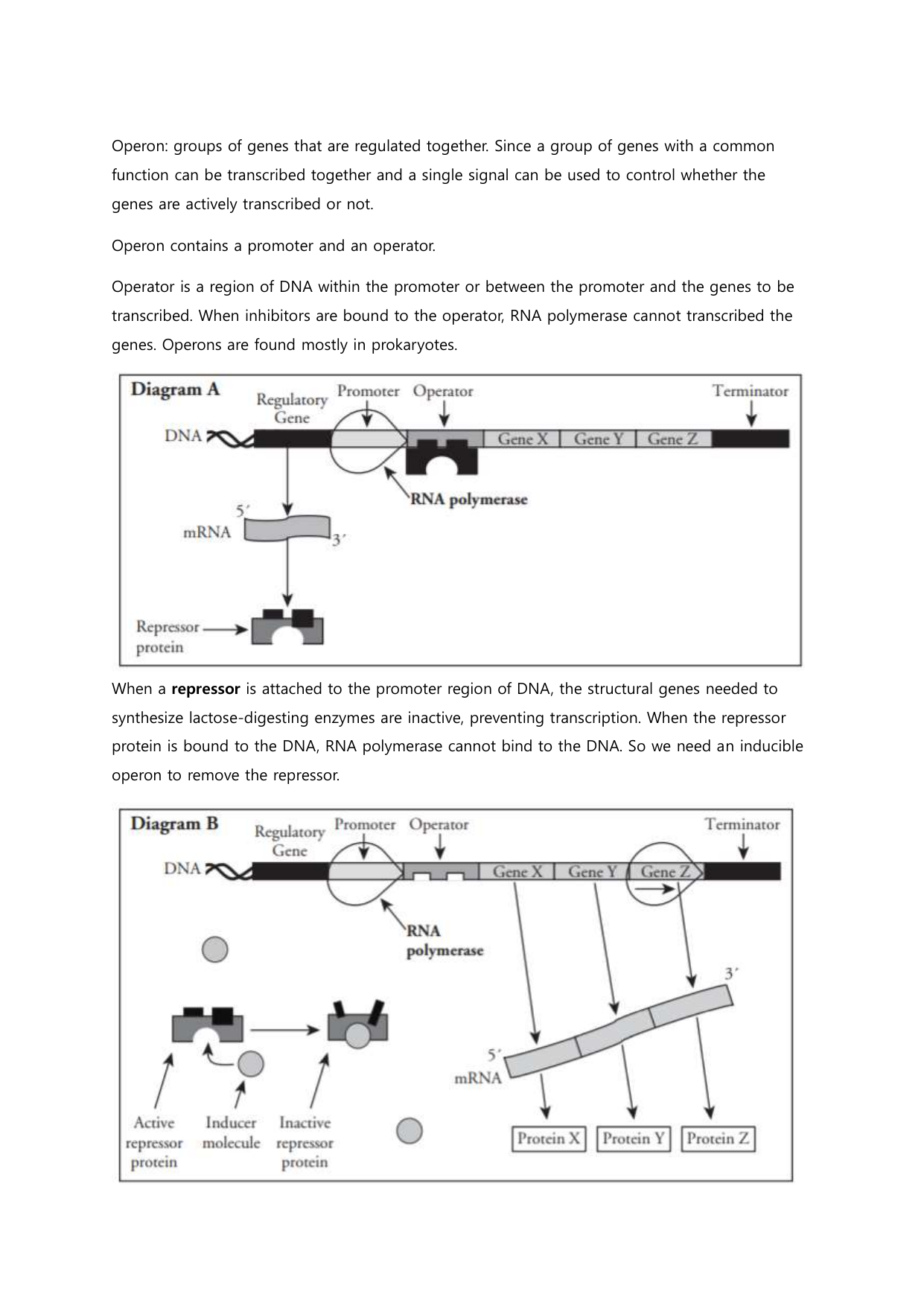
Unlike eukaryotes, where transcription factors shuttle complexly across chromosomes, prokaryotes deploy streamlined systems that enable swift decisions. The *Pogil Control of Gene Expression in Prokaryotes* unit highlights core elements such as promoters—specific DNA sequences where RNA polymerase binds—and operator regions that act as gatekeepers. A classic example is the lac operon in *E.
coli*, a flawless model of regulated expression. “The lac repressor blocks transcription in the absence of lactose,” explains the Pogil framework, “but when lactose is present, it binds to the repressor, enabling RNA polymerase to initiate transcription.” This simple switch exemplifies how prokaryotes conserve resources by expressing genes only when needed. Beyond simple on-off switches, prokaryotes employ layered control systems that integrate environmental signals with internal metabolic status.
Transcriptional regulators fall into several major categories: - **Activators** enhance RNA polymerase binding by binding to operator sequences, such as CAP-cAMP in carbon-starved conditions. - **Repressors** inhibit transcription through direct DNA binding, exemplified by the trp repressor, which halts expression of tryptophan biosynthesis genes when tryptophan levels are high. - **Inducers and substrates**—like lactose for the lac operon or glucose for catabolite repression—modulate regulator activity without direct structural change, enabling dynamic response.
- **Global regulators** such as sigma factors reprogram RNA polymerase recognition across entire operon sets during stress, shifting cellular priorities from growth to survival. These diverse mechanisms form a responsive network rather than isolated switches. The Pogil materials emphasize that prokaryotic cells don’t wait passively—they anticipate and adapt.
For example, under nutrient limitation, cyclic AMP (cAMP) concentrations rise, activating the cAMP receptor protein (CRP). CRP binds upstream activating sequences (UAS) near target genes, recruiting RNA polymerase and boosting transcription of sucrose utilization genes even when primary carbon sources are depleted. Metabolic feedback loops further refine expression control.
When end products accumulate—like isoleucine—repressor proteins bind upstream sequences to suppress their own operon, preventing wasteful synthesis. This negative feedback is a cornerstone of prokaryotic efficiency. “This self-regulating design minimizes metabolic overhead, a triumph of evolutionary selection,” notes the Pogil curriculum, illustrating how survival favors precision at the genetic level.
Another dimension of control centers on translational regulation, occurring after mRNA synthesis. In prokaryotes, where ribosomes diverge rapidly for overlapping genes, regulatory RNAs and protein factors can slow or redirect translation. Riboswitches—NT-segments in mRNA that change structure upon ligand binding—directly couple metabolite levels to protein production.
When the alarm one does not detect abundant amino acids, a riboswitch alters mRNA conformation to halt translation, conserving ATP and nitrogen. This co-transcriptional regulation exemplifies how gene expression is not merely “on” or “off,” but finely tuned to cellular demand. Understanding these processes carries profound implications.
Engineered bacteria now leverage optimized gene circuits for industrial applications—production of biofuels, pharmaceuticals, and enzyme catalysts—relying on the very principles taught through Pogil. Additionally, biomarkers of dysregulated prokaryotic genes inform antibiotic resistance research, where horizontal gene transfer spreads resistance in bacterial populations. “Decoding these regulatory sequences allows targeted intervention,” states a leading microbiologist cited in Pogil case studies, “turning gene control from biological curiosity into therapeutic weapon.” The mastery of gene expression in prokaryotes is not merely academic—it is the foundation of adaptive life at its most fundamental level.
From the operon models of the 1960s to modern CRISPR-based gene editing, the principles reveal a consistent theme: efficiency through regulation. The Pogil framework, grounded in inquiry-driven learning, illuminates this narrative with clarity and depth, showing how a sparse genome orchestrates complexity. In bacteria, where every base pair counts, control is not decoration—it is survival programmed into every instant of expression.
This precision—whether in repressing unnecessary genes, activating emergency pathways, or balancing metabolic loads—mirrors the elegance of biological design. As scientific and industrial applications grow, so does the urgency of understanding prokaryotic gene control, not as an abstract process, but as a blueprint for innovation. With each stacked protein and sequenced operon, nature demonstrates why regulation is the true hallmark of life’s adaptability.
The Operon Model: A Paradigm of Transcriptional Control
The operon stands as the quintessential model for gene regulation in prokaryotes, first elucidated through studies of bacterial metabolism. Central to this architecture are three functional components

Related Post

What Does USNS Mean? Decoding the Abbreviation in Modern Communication

How Jason Benetti’s Net Worth Reflects the Fast-Paced World of High-rolling Entrepreneurship

Is TC Carson Married? The Answer Might Surprise You

